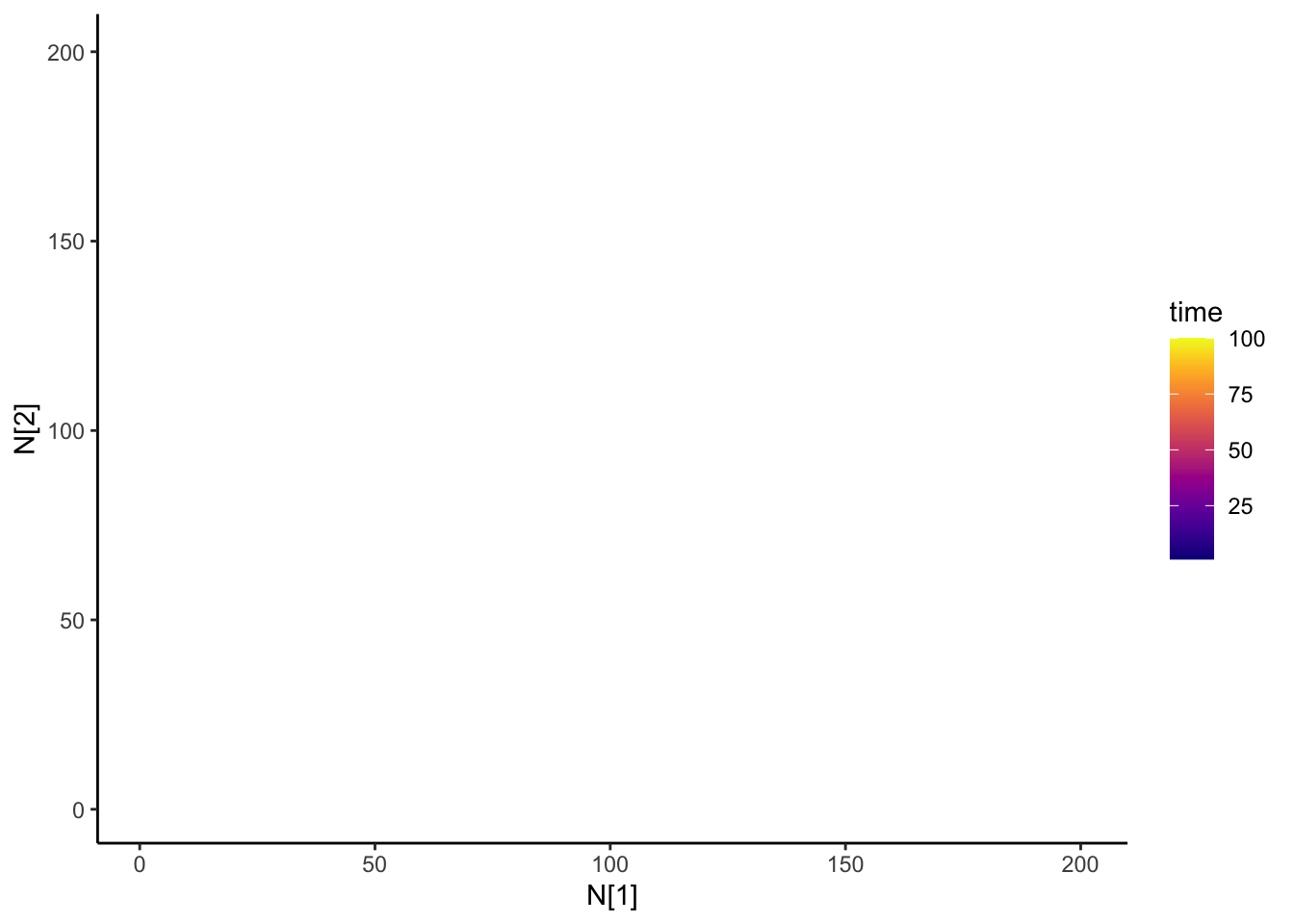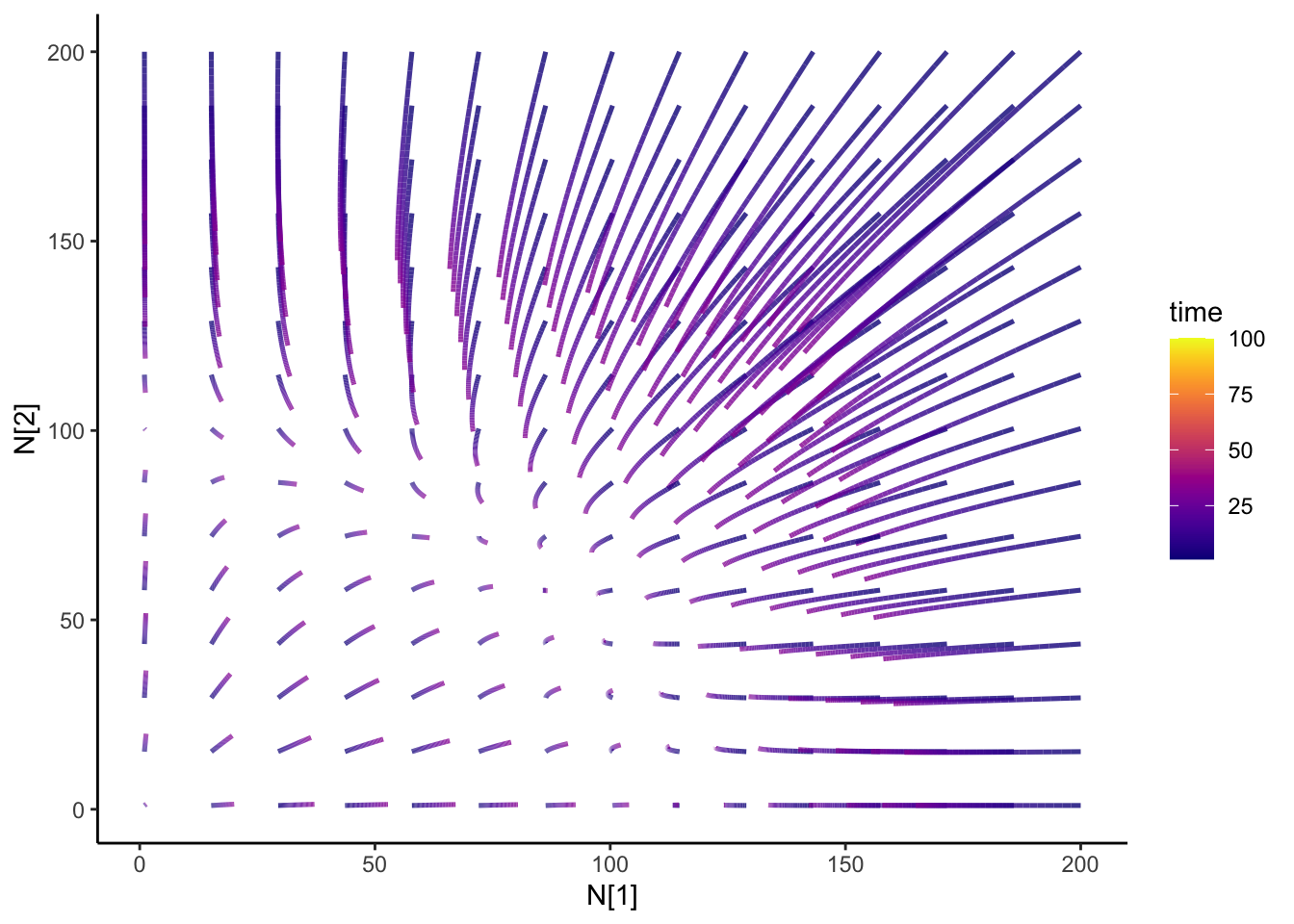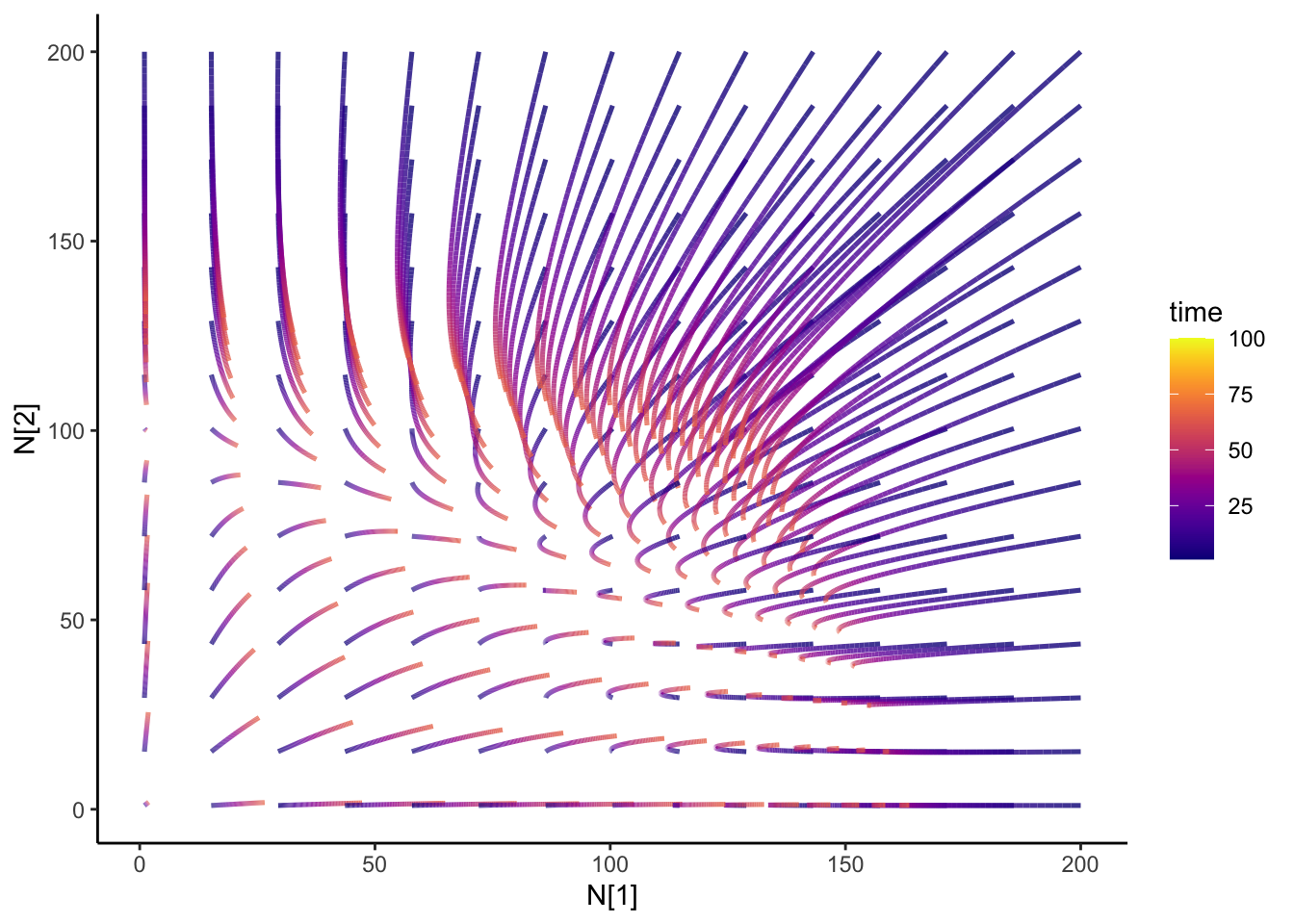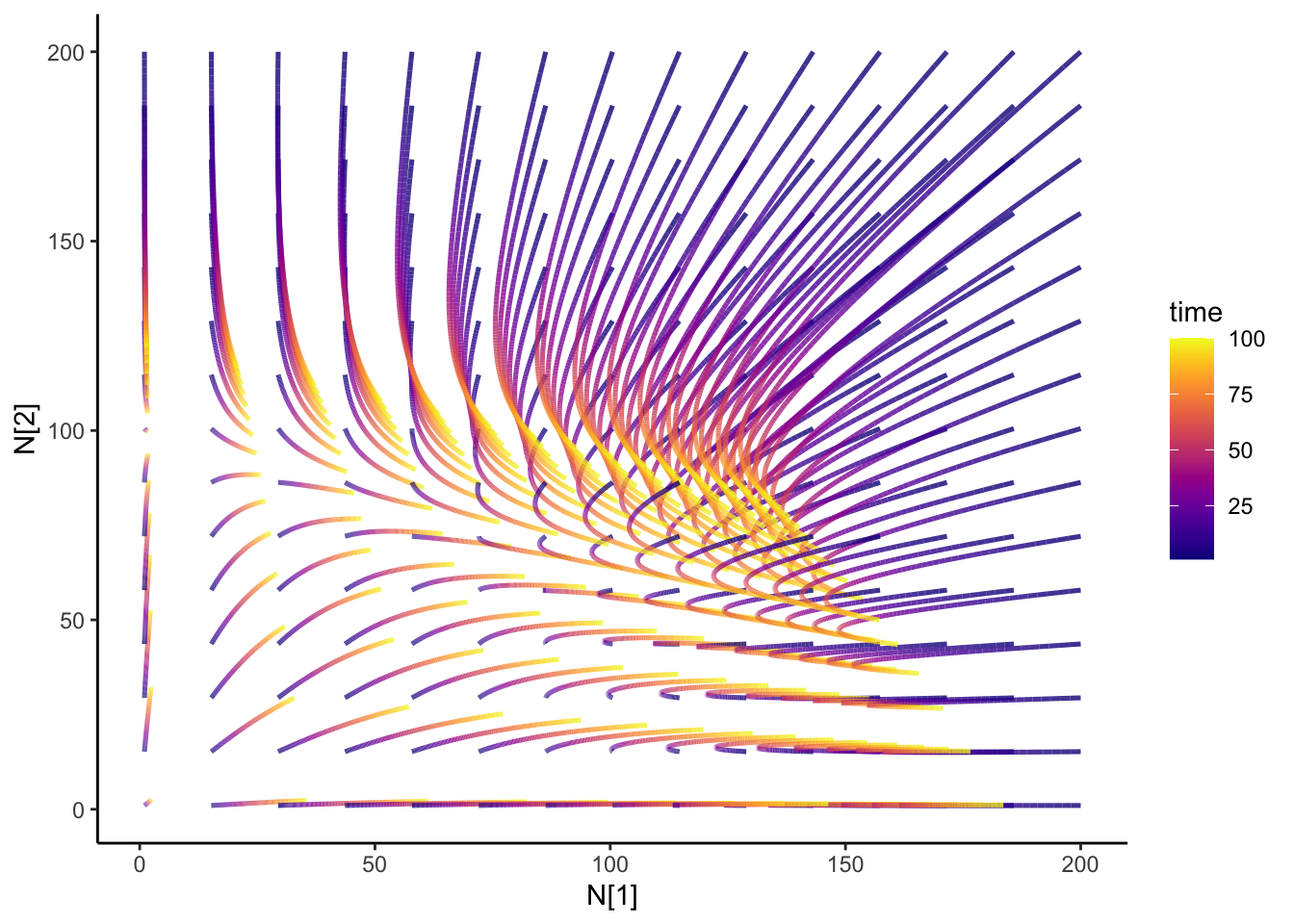library(ggplot2)
library(gganimate)
library(reshape2)
library(viridis)Premise
Ok, so from my last blog post on the Lotka-Volterra competition, I was wondering what would happen if I change my competition parameters over time- how would the dynamics change? Just some code for my future self.
Again, look at my previous post, but I modified the functions:
# c_df is a data.frame containing the competition coefficients above
# N1_0 is the initial population for N1
# N2_0 is the initial population for N2
# timestep is how long we want ot run it for
LV_func <- function(c_df, N1_0, N2_0, timestep) {
# Generate a matrix to keep our records
pop_mat <- matrix(0, nrow = timestep, ncol = 3)
pop_mat[, 1] <- seq(1, timestep)
pop_mat[1, 2] <- N1_0 # initial first population
pop_mat[1, 3] <- N2_0 # initial second population
### Competition coefficient
c_11_v <- c_df["c_11"]
c_12_v <- c_df["c_12"]
c_22_v <- c_df["c_22"]
c_21_v <- c_df["c_21"]
r <- 0.01 # Intrinsic growth rate is same for all species
for (t in seq(1, timestep - 1)) {
c_11 <- c_11_v[t, ]
c_12 <- c_12_v[t, ]
c_22 <- c_22_v[t, ]
c_21 <- c_21_v[t, ]
N1 <- pop_mat[t, 2] # Get the current population sizes
N2 <- pop_mat[t, 3]
N1_change <- r * N1 * (1 - (c_11 * N1 + c_21 * N2))
N2_change <- r * N2 * (1 - (c_22 * N2 + c_12 * N1))
pop_mat[t + 1, 2] <- N1 + N1_change
pop_mat[t + 1, 3] <- N2 + N2_change
}
return(pop_mat)
}timestep <- 100
N12_0 <- expand.grid(
N1 = seq(1, 200, length = 15),
N2 = seq(1, 200, length = 15)
)
c_11_vec <- seq(1e-2, 1e-2 * 0.25, length = timestep)
c_df <- data.frame(
time = seq(1, timestep),
c_11 = c_11_vec,
c_22 = rep(1e-2, timestep),
c_12 = rep(5e-3, timestep),
c_21 = rep(4e-3, timestep)
)N12_full <- do.call(
rbind.data.frame,
apply(N12_0, 1, function(x) {
LV_func(
c_df,
x["N1"],
x["N2"],
timestep = timestep
)
},
simplify = FALSE
)
)
### Add an id
N12_full$id <- rep(seq(1, nrow(N12_0)), each = timestep)
colnames(N12_full) <- c("time", "N1", "N2", "id")isocline_N1 <- function(c_df, timestep) {
ab <- as.numeric(seq(0, 200))
### Competition coefficient
c_11_v <- c_df["c_11"]
c_21_v <- c_df["c_21"]
isocline_change_df <- NULL
for (t in seq(1, timestep)) {
c_11 <- c_11_v[t, ]
c_21 <- c_21_v[t, ]
isocline <- (1 - (c_21 * ab)) / c_11
isocline_change_df[[t]] <- cbind.data.frame(N1 = isocline, N2 = ab, time = as.numeric(t))
}
full <- do.call(rbind, isocline_change_df)
return(full)
}
isocline_N2 <- function(c_df, timestep) {
ab <- as.numeric(seq(0, 200))
### Competition coefficient
c_22_v <- c_df["c_22"]
c_12_v <- c_df["c_12"]
isocline_change_df <- NULL
for (t in seq(1, timestep)) {
c_22 <- c_22_v[t, ]
c_12 <- c_12_v[t, ]
isocline <- (1 - (c_12 * ab)) / c_22
isocline_change_df[[t]] <- cbind.data.frame(N1 = ab, N2 = isocline, time = as.numeric(t))
}
full <- do.call(rbind, isocline_change_df)
return(full)
}isocline1 <- isocline_N1(c_df, timestep)
isocline2 <- isocline_N2(c_df, timestep)That is super-neato!
slate + geom_path(
data = N12_full,
aes(x = N1, y = N2, group = id, color = time), size = 0.9, alpha = 0.8
) +
transition_reveal(time) +
theme_classic() +
scale_color_viridis(option = "plasma") +
ease_aes("cubic-in-out")